Unlocking the Power of Apache Spark: A Comprehensive Guide to Big Data Processing
Introduction: What is Apache Spark?
Apache Spark is a powerful distributed data processing framework for big data workloads. It scales across clusters, enabling efficient batch processing, real-time streaming, machine learning, and graph analytics. Optimised for speed, scalability, and flexibility, Spark leverages in-memory processing to minimise disk I/O and seamlessly handles petabyte-scale data.
Note: Despite its strengths, Spark may not always be the best choice for every scenario. For smaller datasets, its distributed architecture can introduce unnecessary complexity and overhead, making simpler tools like Pandas or Scikit-learn more appropriate. Similarly, Spark is not optimised for ultra-low-latency transactional systems, as it focuses on high-throughput batch and micro-batch processing. While Spark MLlib is useful for large-scale ML training across distributed clusters, many machine learning tasks—particularly those involving smaller datasets or models—benefit from tools optimised for single-node execution and a more straightforward API.
While Spark's capabilities are vast, working with PySpark, its Python API, can sometimes lead to challenges such as long-duration jobs or unexpected failures. These issues often stem from inefficient job logic, suboptimal configurations, or a lack of understanding of Spark's distributed architecture. This guide will walk you through a structured approach to troubleshoot, optimise, and fine-tune PySpark jobs, helping you tackle these challenges and make the most of Spark's powerful ecosystem.
Step 1: Identify the issue
When faced with a PySpark job that takes too long to complete or fails unexpectedly, the first step is to identify the root cause of the issue. Problems can manifest in several ways, including excessively long job durations, frequent job failures, or resource-related errors such as out-of-memory (OOM) exceptions. Understanding these symptoms is crucial for pinpointing the bottlenecks and inefficiencies in your Spark application.
One of the most effective tools for diagnosing such problems is the Spark UI, a built-in web interface that provides detailed insights into job execution. By examining the different tabs in the Spark UI - such as Jobs, Stages, and Executors - you can identify which stages or tasks are taking the longest to complete, whether there are imbalances in task distribution, or if any tasks are failing repeatedly. The UI also allows you to monitor resource usage, shuffle write, disk spillage or review the physical plan respectively its graphical representation.
Understand the logical and the physical plan
As a first step, you should have a look at the logical plan which basically is a Directed Acyclic Graph (DAG, no loops or cycles) and the physical plan (low-level representation derived from the logical plan incl. optimised execution details, such as partitioning, shuffling, and task distribution)

The DAG is Spark's blueprint for executing a job. It divides the job into stages, which are groups of transformations that can be executed in parallel, and tasks, which are the smallest units of execution assigned to individual executors. Each stage in the DAG represents a boundary where Spark performs a shuffle, redistributing data between partitions to meet the requirements of operations like groupBy or join. These shuffle operations are often expensive and can significantly impact performance if not managed properly.
By inspecting the DAG, either through the Spark UI (SQL tab) or programmatically, you can identify the key stages of your job and the transformations that contribute to each stage. Look for patterns such as a large number of stages, an unusually high shuffle size, or tasks that are disproportionately slow. These are often indicators of issues like data skew, unnecessary transformations, or suboptimal partitioning.
Use the explain() Function on any dataframe during development to obtain a detailed view of the logical (via extended mode) and the physical plan. I suggest that when you type code you already think about how the corresponding physical plan could look like.
Step 2: Minimise data shuffle
A shuffle in Apache Spark occurs when data needs to be redistributed across partitions, typically in wide transformations like groupBy(), join(), orderBy(), or repartition().
Wide vs. Narrow Transformations: In Spark, transformations are classified as narrow or wide based on how they process data across partitions. Narrow transformations like map, filter, and select operate within individual partitions without requiring data movement, making them efficient since they do not trigger costly shuffling. In contrast, wide transformations like groupBy, join, and orderBy require redistributing data across partitions, causing shuffle operations that involve network communication and disk I/O, which can impact performance. To optimise Spark applications, it is best to minimise wide transformations and use partitioning strategies when necessary to reduce shuffling and improve efficiency.
Since shuffling involves network transfers and disk writes, it can be costly, leading to higher latency, increased memory usage, and potential job slowdowns, especially if data is skewed. To optimise performance, strategies like broadcast joins, partition pruning, and careful partitioning should be used to minimise unnecessary shuffling.
a. Use broadcast join
A broadcast join is an optimisation technique in Apache Spark that improves the performance of joins involving a small dataset and a large dataset. Instead of shuffling data across worker nodes, Spark broadcasts the smaller dataset to all executors, allowing each node to perform the join locally. This significantly reduces network I/O and shuffle costs, making the join operation much faster. Broadcast joins are particularly useful when joining a large DataFrame with a small lookup table that fits in memory. Spark automatically determines when to use a broadcast join based on the size of the smaller DataFrame (default threshold: 10 MB), but you can explicitly enforce it using broadcast() from pyspark.sql.functions. Using broadcast joins prevents expensive shuffles, reduces memory pressure, and enhances query performance, making it a best practice for optimising joins when dealing with small reference tables.
# Use broadcast join
result_df = large_df.join(broadcast(small_df), on="state", how="inner")
c. Window functions: Using window functions can be an effective way to reduce shuffle operations in Spark. While a groupBy typically requires a shuffle to aggregate data across partitions, window functions can often avoid this overhead if the data is already partitioned correctly. However, when sorting is required (through an orderBy clause), it may still trigger a shuffle. Understanding the interaction between sorting and shuffling is key to optimising performance.
A global groupBy always requires a shuffle, as Spark needs to collect and group data by key across partitions, but it does not inherently require sorting. In contrast, a window function with partitionBy only triggers a shuffle if the data is not already partitioned by the specified key. If an orderBy is also used within the window, sorting is required within each partition, which usually results in a shuffle.
For specific window operations, a sum over a window behaves similarly, only requiring a shuffle if the partitionBy keys do not align with the existing partitions. Sorting is not needed unless orderBy is explicitly used. On the other hand, retrieving the first value in a window follows the same partitioning rules as a sum but will require sorting if an orderBy is present, leading to a shuffle.
c. Combine Small Files: Small files can cause excessive shuffle due to metadata overhead. Consolidate them during writes. You can for instance use coalesce to combine or use the following statement:
python df.write.option("maxRecordsPerFile", 100000).parquet("output_path")
Step 3: Optimise dataset partitions
a. Options
If shuffling is unavoidable, you must decide how to manage it efficiently, as it typically involves repartitioning the data across nodes.
The best option for partitioning (whether to intervene or not) largely depends on your specific use case, the dataset size, and the structure of your pipeline. Let's break down each option and when it's most appropriate:
- Option A: Let Spark handle partitioning automatically - recommended for small datasets
- Option B: Repartition within the current job - this is the safest and most flexible choice for single-stage jobs with shuffle-heavy transformations, potential data skew or imbalanced partitions, as it allows fine-tuning the number of partitions explicitly.
- Option C: Repartition when writing files (upstream partition optimization which can be reused across multiple downstream jobs) - most efficient for multi-stage jobs with reusable datasets, as it avoids redundant shuffling in downstream jobs.
If you're working with a large datasets (applies to options B. or C.), you might need more than 200 partitions to ensure each partition is small enough to fit in memory.
# Adjust shuffle partitions
spark.conf.set("spark.sql.shuffle.partitions", "500")You can also explicitly repartition your dataset to maintain a consistent partitioning scheme across multiple operations, reducing unnecessary shuffling. Without this, each wide transformation (e.g., join, groupBy) may trigger its own shuffle. Shuffling is required when a dataframe cannot be broadcasted, as join keys must be co-located in the same partition for proper mapping.
df = df.repartition(100, "key") # Explicitly use 100 partitions
b. Data Skew
Repartitioning can distribute data across multiple or a specified number of partitions, but it does not guarantee even distribution, potentially leading to data skew, where some executors are overloaded while others process minimal data.
- If your dataset contains a numerically ordered key, using
repartitionByRange()allows Spark to dynamically calculate ranges, helping to distribute data more evenly, especially for keys like numeric IDs, timestamps, or sorted categories. - If no such key exists, salting with an initial factor of 5 or 10 can introduce randomness into partitioning, reducing skew and ensuring a more balanced workload across executors.
from pyspark.sql.functions import col, concat, lit, rand, split, sum as _sum# Step 1: Read the datadf = spark.read.parquet("data.parquet")# Step 2: Add a "salt" column# Generate a random number (or range of numbers) to create a salted keynumsalts = 10 # Choose a salt range (e.g., 10 salts)df = df.withColumn("saltedkey", concat(col("key"), lit(""), (rand() * numsalts).cast("int")))# Step 3: Repartition by the salted keydf = df.repartition(100, "saltedkey")# Step 4: Perform the groupBy on the salted keysaltedresult = df.groupBy("saltedkey").agg({"value": "sum"})# Step 5: Remove the salt and aggregate by the original keyfinalresult = saltedresult.withColumn("key", split(col("saltedkey"), "")) \ .groupBy("key").agg(_sum("sum(value)").alias("sumvalue"))c. Disk spillage
Disk spillage during shuffle occurs when the data being shuffled exceeds the available memory, causing Spark to write the excess data to disk, which can significantly slow down the processing. Disk spillage frequently happens during wide transformations or large joins; below are some remedies to address this issue:
-
Increase Memory Allocation for Executors: Allocate more memory to each executor to reduce the likelihood of spilling.
python from pyspark.sql import SparkSession
spark = SparkSession.builder \
.appName("OptimizeSpillage") \
.config("spark.executor.memory", "8g") \
.config("spark.executor.memoryOverhead", "2g") \
.getOrCreate() -
Optimize partitioning to reduce data skew see measures at the beginning of this chapter
-
Persist Data Strategically: Cache or persist intermediate results in memory to reduce re-computation and spillage.
df_cached = df.persist()
df_cached.count() # Trigger the cache
Note: Disk spillage is not limited to shuffling but can occur in other stages of data processing as well, such as during aggregations, joins, or any operation that requires holding a large amount of intermediate data in memory. Essentially, any time the available memory is exceeded, spillage can occur, regardless of the specific operation being performed
Step 4: Tune Spark configuration
When tuning Spark configurations, different types of jobs require different optimizations:
a. Worker-Intensive Jobs
Worker-intensive jobs process large datasets (10 GB to 1 TB+) with heavy computation and data shuffling. Operations like join(), groupBy(), and complex UDFs rely on distributed execution across worker nodes. Large partitions enhance parallelism but can overload executors if not tuned properly. Shuffling-intensive operations like sorting and aggregation require careful memory management to avoid excessive disk usage.
Tuning Options:
- Increase executor memory to handle large workloads efficiently:
spark.conf.set("spark.executor.memory", "8g")
spark.conf.set("spark.executor.memoryOverhead", "2g") - Optimize shuffle partitions to balance load across executors:
spark.conf.set("spark.sql.shuffle.partitions", "500") # Adjust based on workload - Use efficient partitioning to minimize data movement:
df = df.repartition(200, "key_column") # Ensures partitions are aligned to keys
b. Driver-Intensive Jobs
Driver-intensive jobs require significant coordination, especially for collecting large results, performing aggregations, or broadcasting data. These jobs are common in workloads involving frequent collect(), complex SQL queries, or iterative computations where too much metadata processing burdens the driver.
Tuning Options:
- Increase driver memory to prevent out-of-memory errors:
spark.conf.set("spark.driver.memory", "8g") - Avoid
collect()and use actions that minimize data transfer to the driver:df.agg({"column": "sum"}).show() # Efficient
df.count() # Efficient, as only small numbers are returned
df.collect() # Inefficient, should be avoided for large datasets
c. Jobs Intensive on Both Driver and Workers
These jobs handle large-scale data (100 GB+) while requiring significant driver coordination. Workloads like iterative machine learning (fit() and transform() in Spark ML), complex queries with multiple joins and aggregations, or broadcasting large variables fall into this category. Optimizing memory allocation and partitioning is key to balancing execution across driver and worker nodes.
Tuning Options:
-
Balance driver and executor memory:
spark.conf.set("spark.driver.memory", "6g")
spark.conf.set("spark.executor.memory", "10g") -
Optimize parallelism by tuning partition sizes and shuffle behavior:
spark.conf.set("spark.sql.shuffle.partitions", "400") # Adjust based on workload
d. Be adaptive
Enabling dynamic resource allocation allows Spark to scale the number of executors up or down based on workload demands, optimizing resource usage and cost efficiency.
Tuning Options:
-
Enable dynamic allocation in Spark to scale executors as required:
spark.conf.set("spark.dynamicAllocation.enabled", "true") spark.conf.set("spark.dynamicAllocation.minExecutors", "2") spark.conf.set("spark.dynamicAllocation.maxExecutors", "10") -
Enable Adaptive Query Execution (AQE), introduced in Spark 3.0, which reduces shuffle overhead and improves query efficiency, making Spark execution more adaptive to real-world data distribution:
spark.conf.set("spark.sql.adaptive.enabled", "true")
Step 5: Optimise the code
Optimising code in PySpark is crucial for enhancing performance and efficiency. This step delves into various techniques and best practices for improving PySpark code, including the judicious use of unionByName(), predicate pushdown, caching strategies, leveraging the Spark API, incremental tuning, and handling complex jobs. Actually, you might want to consider these optimizations before configuration tuning (as discussed in the previous step), as they can have a substantial impact on the required configuration settings, such as partition shuffle size.
a. Why You Should Use unionByName() Sparingly in PySpark
In PySpark, unionByName() aligns columns by name rather than position, but excessive use inflates the query plan, leading to performance issues. Since Spark follows a lazy execution model for fault tolerance and resilience, transformations are not executed immediately but recorded in a logical execution plan. Each unionByName() call expands the query lineage, duplicating all prior transformations of the combined DataFrames. Unlike Python, where operations modify objects in memory instantly, Spark's DAG execution model retains transformations until an action (e.g., count(), collect(), write()) triggers execution. Overusing unionByName() before an action forces Spark to recompute dependencies, causing redundant computation and inefficiency.
We have two examples: one using unionByName() and another without it. Replacing unionByName() with a when clause not only makes the code more concise but also optimizes the physical execution plan, reducing unnecessary computation. This improvement significantly lowers the runtime, cutting it from 30 to 20 minutes in our example, which is around a 30% improvement.
df_A = df.filter(F.col("TYPE") == "A").withColumn("SAMPLES", samples_A_udf())
df_B = df.filter(F.col("TYPE") != "A").withColumn("SAMPLES", samples_B_udf())
df = df_A.unionByName(df_B)
df = (df.select("*", F.posexplode("SAMPLES").alias("POS", "GROWTH"))
.withColumn("AMOUNT", F.col("GROWTH") * F.col("AMOUNT")))Above code results in a physical plan that duplicates the transformation steps within this one stage (you can visually detect it based on the recurring text pattern of the physical plan).

df = (df.withColumn("SAMPLES", F.when(F.col("TYPE") != "A", samples_B_udf()).otherwise(samples_A_udf()))
.select("*", F.posexplode("SAMPLES").alias("POS", "GROWTH"))
.withColumn("AMOUNT", F.col("GROWTH") * F.col("AMOUNT")))
The improved code using the when clause (physical plan below) eliminates the duplication of transformation steps, making the execution plan more efficient.
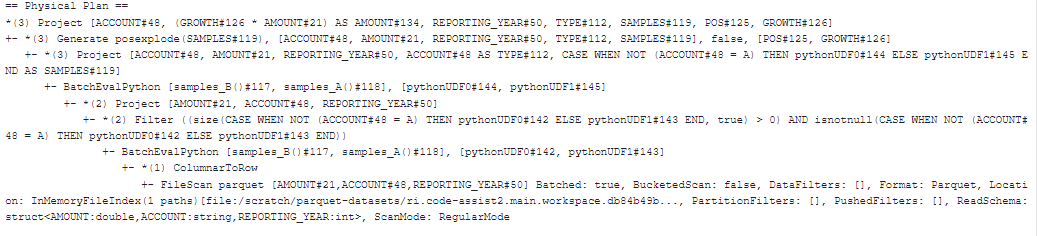
The output DataFrame is the same in both cases. It generates a new column called SAMPLES based on the TYPE column using different UDFs. After creating the SAMPLES column, the posexplode() function is used to explode the SAMPLES array, creating a new row for each element in the array. Finally, the AMOUNT column is updated by multiplying it with the exploded GROWTH values.
A possible application for this process could be in financial data analysis. For instance, you might have a dataset containing transaction records with different types of financial instruments and want to generate samples based on the type of instrument for further analysis or simulations.
b. Pushing down filters
Predicate pushdown is an optimization technique that allows Spark to filter data at the source (e.g., while reading from Parquet) instead of loading unnecessary rows into memory. This significantly reduces I/O, memory usage, and processing time by ensuring only relevant data is read. Predicate pushdown works automatically for columnar formats like Parquet and ORC, where Spark can push filters to the storage layer. However, it may not be applied when using complex expressions, UDFs, or formats like JSON and CSV, which require full data scans. To benefit from pushdown, always apply filter() or where() as early as possible in your query plan, ensuring Spark reads only the necessary subset of data instead of filtering after loading everything into memory.
Spark enables predicate pushdown by default, but you can ensure it's active:spark.conf.set("spark.sql.parquet.filterPushdown", "true")
Example code:
df = spark.read.parquet("path/to/output").filter("region = 'US'")
c. When to Cache and When Not To
In Spark, if a DataFrame is used multiple times, Spark recomputes all prior transformations each time it is referenced. This can cause unnecessary shuffling and increased execution time, especially for transformations like groupBy(), join(), or repartition(). Caching helps avoid this recomputation by storing the DataFrame in memory or disk, allowing Spark to directly access the cached data instead of re-executing previous transformations.
However, caching comes at a cost - it consumes memory or disk space, depending on the persistence level. If memory is insufficient, caching large datasets may cause garbage collection issues or force data eviction, reducing efficiency. Therefore, caching should only be used when the same DataFrame is reused multiple times in a job, the cached dataset is expensive to recompute (e.g., involving wide transformations), and the cluster has enough storage resources to hold the cached data without impacting other tasks.
For large datasets or memory-limited environments, DISK_ONLY is often preferred as a persistence level to avoid memory pressure and unpredictable eviction, ensuring stability.
Caching is lazy - it does not take effect until an action (e.g., count()) is executed. To ensure caching is applied, trigger it with an action:df.persist("DISK_ONLY") df.count() # Forces caching to take effect
While caching can reduce recomputation and optimize performance, it should be used strategically to balance efficiency and resource consumption.
d. Leverage the Spark API whenever possible
In PySpark, you should always use built-in functions instead of user defined functions (UDFs) whenever possible, as they are optimized for distributed execution and leverage Spark's Catalyst optimizer. UDFs, on the other hand, introduce performance overhead because they run row-by-row in Python (via Py4J), breaking Spark's native optimization pipeline and slowing down execution, especially on large datasets. For example, if your DataFrame requires a column with a sequence of integers, you might consider generating it manually.
# Define the UDF for sequence generation
def generate_sequence(start, end, step=1):
if start is None or end is None or step == 0:
return []
return list(range(start, end + 1, step))
# Register UDF with appropriate return type
sequence_udf = udf(generate_sequence, ArrayType(IntegerType()))
However, PySpark provides a built-in function, f.sequence(), that achieves this efficiently. Always check the latest API documentation before implementing custom solutions to avoid unnecessary complexity and performance overhead.
e. One change at a time
When tuning PySpark performance, it's essential to adjust parameters or code incrementally rather than making multiple changes simultaneously. This approach helps isolate the impact of each optimization, making it easier to identify what enhances performance and what doesn't. Begin with small adjustments, such as optimizing partitioning, memory allocation, or shuffle settings, and test iteratively to avoid unintended performance regressions. This method ensures a more controlled and effective tuning process.
f. When a single job becomes inefficient
Sometimes, jobs attempt to handle too many transformations in one execution, making pre-partitioning ineffective as key distributions shift across multiple wide transformations. This can lead to imbalanced workloads, excessive shuffling, and inefficient execution. In such cases, two possible solutions can help:
Checkpointing: Using checkpointing splits the DAG, preventing Spark from holding the entire lineage in memory. While checkpointing introduces some memory and metadata overhead (while saving the intermediate state of the data processing pipeline to a reliable storage system as S3), it improves fault tolerance and simplifies debugging.
Splitting jobs: Splitting the job into two sequential executions can sometimes be faster and more efficient than running a single, overloaded transformation. Although this approach requires careful resource management and may introduce data transfer overhead, it can significantly reduce execution time by isolating shuffle-heavy stages.
Choosing between checkpointing and job splitting depends on workload size, shuffle intensity, and resource constraints, ensuring optimized performance and better scalability.
Step 6: Accelerate disk I/O
Efficient read and write speeds are crucial for achieving optimal performance in any data-driven application. By adjusting the Spark settings to match the specific characteristics of your job's data, you can ensure that your data flows smoothly and quickly through the pipeline. The following configuration settings have a direct impact on reading and writing performance:
a. parquet.block.size
The block size parameter in Parquet files determines the amount of data written into a single block, impacting both read and write performance. Larger blocks (default ~128 MB) enhance read efficiency by reducing the number of blocks and metadata, but slightly increase write time. Smaller blocks increase metadata overhead and random I/O, generally leading to slower reads. For best performance, use a block size of 256 MB or higher for large analytic workloads, while smaller block sizes may suit small files.ctx.spark_session.conf.set("parquet.block.size", 256*2**20)
b.spark.sql.files.maxPartitionBytes
The spark.sql.files.maxPartitionBytes parameter controls the maximum data size per partition during file reads in Spark, impacting how data is split for parallel processing. By default, it's set to 128 MB.
Larger partition sizes reduce the number of tasks, lowering scheduling overhead and improving performance on powerful clusters but can increase executor memory usage. Smaller partitions enable greater parallelism and are better for memory-constrained nodes, though they may increase task scheduling overhead. Please note that this setting doesn't affect writes but influences how files are split during reads.
To optimize performance, align the partition size with executor memory and workload needs - i.e. use larger sizes (e.g., 256 - 512 MB) for large files and analytics, while smaller values suit memory-limited environments or small datasets.
ctx.spark_session.conf.set("spark.sql.files.maxPartitionBytes", "256mb")
c. Partition Your Data
Partitioning is also important for accelerating I/O due to its ability to organize data into smaller, logical chunks, which improves read and write performance by enabling partition pruning and parallelism.
When writing data, partitioning by frequently queried columns (e.g., date, region) improves query performance, while avoiding high-cardinality columns (e.g., user_id) prevents excessive small files. Too many small files lead to high task overhead and inefficient parallelism, so using repartition() or coalesce() helps optimize file distribution. Aim for file sizes between 128-256 MB for better read performance and adjust the number of partitions accordingly to balance workload efficiency.
# Partitioning by year, month, and day
df.write.partitionBy("year", "month", "day").parquet("path/to/output")# Repartition increases partitionsdf.repartition(100).write.parquet("path/to/output")
# Coalesce reduces the number of partitions (best for frequently queried columns, avoiding the scanning of numerous small partitions)
df.coalesce(10).write.parquet("path/to/output")d. Optimize Shuffle Operations
Shuffles during writing can cause excessive disk I/O, so optimizing them is crucial for improving performance. To prevent the creation of too many small partitions, adjust the shuffle partitions based on the dataset size. As mentioned in the configuration tuning section, the shuffle partitions parameter is key for jobs with heavy loads on workers. The default setting (spark.sql.shuffle.partitions = 200) works for moderate workloads, but for large datasets, increasing this value can enhance parallelism, while decreasing it for smaller datasets can reduce overhead.
# Adjust shuffle partitions
spark.conf.set("spark.sql.shuffle.partitions", "500")Conclusion
Apache Spark is a powerful framework for big data processing, offering scalability, speed, and flexibility. However, optimizing PySpark jobs is crucial to fully leverage its potential, especially when dealing with large datasets and complex transformations. This guide has outlined key steps to diagnose performance bottlenecks, minimize data shuffling, optimize partitioning, tune Spark configurations, and refine PySpark code for efficiency. Following the key takeaways:
- Diagnose Issues Early – Use the Spark UI and
explain()function to analyze job execution and identify inefficiencies in logical and physical plans. - Minimize Data Shuffle – Avoid excessive shuffling by leveraging broadcast joins, window functions, and effective partitioning strategies.
- Optimize Dataset Partitioning – Choose appropriate partitioning strategies (automatic, in-job, or upstream) based on workload requirements to balance performance and resource utilization.
- Tune Spark Configuration – Adjust memory allocation, shuffle partitions, and enable adaptive query execution (AQE) to optimize Spark’s resource usage.
- Refine PySpark Code – Reduce dependency on
unionByName(), leverage built-in functions instead of UDFs, and use caching strategically to minimize unnecessary recomputation. - Accelerate Disk I/O – Optimize file partitioning, block size, and shuffle settings to improve read and write efficiency.
In real-world applications, tuning PySpark is an iterative process. Continuous monitoring, experimentation with configurations, and iterative code optimizations will yield the best results. From my perspective encountering performance challenges or job failures should be seen as an opportunity to sharpen your Spark expertise. Each troubleshooting experience enhances your ability to design better workflows, write more efficient code, and configure Spark for optimal performance. With a combination of curiosity, attention to detail, and adherence to best practices, you can master PySpark and unlock its full potential for tackling big data workloads.
Now it's your turn to put these principles into action. Use what you have learned here to analyze your own PySpark applications, experiment with optimizations, and continue exploring the many ways Spark can drive your data projects forward.
Alberto Desiderio is deeply passionate about data analytics, particularly in the contexts of financial investment, sports, and geospatial data. He thrives on projects that blend these domains, uncovering insights that drive smarter financial decisions, optimise athletic performance, or reveal geographic trends.



